Effortlessly Merging 1,000s of Raw Data Files with Stata
I frequently have to consolidate 100s or 1,000s of raw data files into Stata, that are potentially stored in numerous and potentially unknown folders and subfolders, and have developed a workflow that I think is useful. This approach means I don’t need to know file names or paths, and can instead assign search parameters that determine which files get tagged for processing. I’ve used this approach when constructing a financial flows panel database for India from numerous state-level bank deposits spreadsheets, as well as manipulating GCM output that was chopped up and spit out into thousands of files using an Python/ArcPy workflow. I frequently encounter these setups, and if you do too then this tutorial is probably relevant. All the files needed to work through the following example are on github.
Philosophy
This merge procedure satisfies two key requirements:
- Agile – hard-coding is either minimized or non-existent, which means you can update folder contents, including the addition of new subfolders, and the script will automatically adjust. Agility would not be achieved, for example, by using “medium-coding” solutions (somewhere in the middle of soft and hard) like for loops with fixed end points, since changes to the range do not necessarily alter the for loop structure. To be concrete, assume someone from the House Intelligence Committee handed you files that spanned “WiretappingRawData_32.csv” to “WiretappingRawData_56.csv.” You could write a for loop spanning 32 to 56, but this does nothing when files *_57 onwards are added.
- Versatile – can easily be adapted to a range of data formats and processing requirements. The major code chunks below can easily accommodate CSVs, XLSXs, binary files, etc., and slots into longer Stata workflows.
Data Context
Let’s start with this example, which includes some simplifying elements that we can revisit later. You need to import daily temperature data for several climate models for multiple climate experiments (e.g., representative concentration pathways) for a single year, which quickly puts us in the realm of 10,000s of files. The directory tree may look like this:
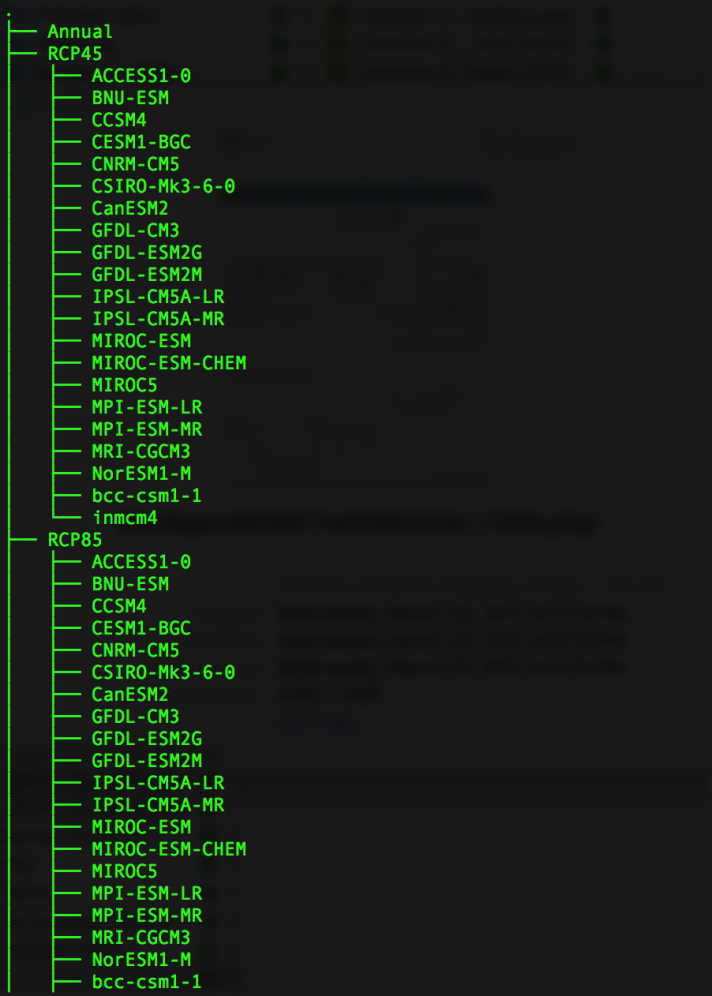 where RCP45 and RCP85 respectively refer to output from the RCP 4.5 and 8.5 experiments. All of the 20+ subfolders (ACCESS, BNU, MIROC, etc.) have names that are unknowable in advance, and the lack of a predictable naming scheme necessitates an ‘agile’ approach which organically discovers all the paths. In each subfolder are numerous model-specific files, each containing data for an individual date, like these:
where RCP45 and RCP85 respectively refer to output from the RCP 4.5 and 8.5 experiments. All of the 20+ subfolders (ACCESS, BNU, MIROC, etc.) have names that are unknowable in advance, and the lack of a predictable naming scheme necessitates an ‘agile’ approach which organically discovers all the paths. In each subfolder are numerous model-specific files, each containing data for an individual date, like these:
 You first need to install ashell which allows us to handily capture and manipulate shell output.
You first need to install ashell which allows us to handily capture and manipulate shell output.
We’ll use find which is a basic shell command that can be accessed by Stata.
ashell find "${cmip5Base}" -maxdepth 1 -mindepth 1 -type d
This command uses my global ${cmip5Base} folder as the root directory, searches 1 level deep (no more, no less, as dictated by the maxdepth and mindepth parameters) and only searches for paths, denoted by the “-type d” suffix referring to directories, not files.
Ashell will print out the series of paths or files that satisfy the find search, and uses this very helpful indexing system which can be managed with a for loop, where each search return is numerically identified as 1 through `r(no)’, which is the last file. Let’s assume we just want to list the directory contents, and so running the following loop will print out all those paths.
forvalues j = 1/`r(no)' {
loc currentDir "`r(o`j')'"
di "Current Dir = `currentDir'"
}
Now we want to access the files in each of the j folders. They are accessible as r-class macros, starting with `r(o1)’ and ending at some J = no. We issue a similar find command, but now stipulate some file parameters.
ashell find "`currentDir'" -maxdepth 1 -name "*.csv"
Instead of the -type option, we’re now using -name with a wildcard to capture only comma-separated files. Keeping maxdepth to 1 restricts the search to only those files stored in `currentDir.’
This generates another r-class counter spanning ``r(o1)’ to r(no)’, which can be handled via a new and differently indexed for loop across i. Consider this:
forval i = 1/`r(no)' {
di "r(o`i')"
insheet using "`r(o`i')'", comma clear
}
Since our target raw data files are CSVs, we need to use the insheet command instead of import excel.
I then rely on Stata’s extremely helpful regular expressions operators to extract important details from the filename. In order to do this, you would know ex ante the file naming convention and how to access each segment. As an example, consider a file named
tasmax_day_BCSD_rcp85_r1i1p1_CanESM2_2050-01-01_1991District__Power_3.csv
which tells me which variable (tasmax_day), which experiment (rcpXX), which model (CanESM2), which date (2050-01-01), and which polynomial degree (Power_3) this data is taken from. Ideally, all these pieces would be included in the data itself, but let’s assume they’re not. One way of using regular expressions to strip out those identifiers look something like this, though obviously there are numerous ways you can tackle this:
* Recover basics from filename, presumably more accurate than if sourced from directory name *
gen polynomialOrder = regexs(2) if regexm("`fn'", "(Power_)([0-9])(.csv)")
gen modelFamily = regexs(2) if regexm("`fn'", "(r1i1p1_)([a-zA-Z0-9-]*)(_2050)")
gen experiment = regexs(2) if regexm("`fn'", "(BCSD_)([a-zA-Z0-9-]*)(_r1)")
gen year = regexs(1) if regexm(dateFixed, "([0-9][0-9][0-9][0-9])(-)")
gen month = regexs(2) if regexm(dateFixed, "(-)([0-9][0-9])(-)")
gen day = regexs(4) if regexm(dateFixed, "(-)([0-9][0-9])(-)([0-9][0-9])")
destring, replace
gen date = mdy(month, day, year)
format date %td
Let’s say those variables are sufficient to uniquely identify our observations, since we want to merge this particular file with 1000s of others and not worry about those gruesome _merge == 5 errors. This particular spreadsheet is then saved as a temporary file, which can then be seamlessly merged with all others. As an example, we can do this:
tempfile temp`i'
save `temp`i''
di "Just finished saving file `i'"
ashell find "`currentDir'" -maxdepth 1 -name "*.csv"
}
use `temp1', clear
forval j = 2/`r(no)' {
merge 1:1 dist_code date modelFamily experiment using `temp`j'', update replace
tab _merge
drop _merge
}
The reason for issuing a second instance of ashell before entering the next iteration of the i for loop, is because of a quirk which leads the r(o1)…r(no) local index values to be erased when some data processing command is performed. Once the merge is complete, you can then save your master file as a *.dta.
While the Stata .do I’m sharing explicitly identifies the parent folder, you can change the -maxdepth parameters to search across multiple sets of children paths, with the ability to save all sub-parent folder contents as a separate *.dta. You can then repeat the procedure abov, across your individual *.dta’s, to merge up into a master *.dta. The reason you might wish to adopt an iterative approach is that each subroutine requires lengthy processing time, and you can avoid duplicating work by saving results separately.
I’d love to hear from you if you found this helpful, or have suggestions for tailoring this to specific workflow requirements you frequently find yourself having to plough through.